Beta-glucuronidase
| beta-glucuronidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Glucuronidase Homotetramer (assumed biological unit)[1] | |||||||||
| Identifiers | |||||||||
| EC number | 3.2.1.31 | ||||||||
| CAS number | 9001-45-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| glucuronidase, beta | |
|---|---|
 Beta-glucuronidase asymmetric unit showing active site residues Glu451, Tyr504, and Glu540, along with the potentially supporting Asn450 residue[1] | |
| Identifiers | |
| Symbol | GUSB |
| Entrez | 2990 |
| HUGO | 4696 |
| OMIM | 611499 |
| RefSeq | NM_000181 |
| UniProt | P08236 |
| Other data | |
| EC number | 3.2.1.31 |
| Locus | Chr. 7 q11.21 |
Beta-glucuronidases are members of the glycosidase family of enzymes that catalyze breakdown of complex carbohydrates.[2] Human β-glucuronidase is a type of glucuronidase (a member of glycosidase Family 2) that catalyzes hydrolysis of β-D-glucuronic acid residues from the non-reducing end of mucopolysaccharides (also referred to as glycosaminoglycans) such as heparan sulfate.[2][3][4] Human β-glucuronidase is located in the lysosome.[5] In the gut, brush border β-glucuronidase converts conjugated bilirubin to the unconjugated form for reabsorption. Beta-glucuronidase is also present in breast milk, which contributes to neonatal jaundice. The protein is encoded by the GUSB gene.[6][7]
Contents
1 Structure
2 Mechanism of catalysis
3 Sly syndrome
4 Molecular applications: use as a reporter gene
5 See also
6 References
7 Further reading
8 External links
Structure
Human β-glucuronidase is synthesized as an 80 kDa monomer (653 amino acids) before proteolysis removes 18 amino acids from the C-terminal end to form a 78 kDa monomer.[8][9]
Beta-glucuronidase exists as a 332 kDa homotetramer.[10] Beta-glucuronidase contains several notable structural formations, including a type of beta barrel known as a jelly roll barrel and a TIM barrel.[1]
Mechanism of catalysis
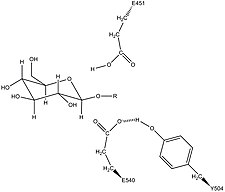
Human β-glucuronidase is homologous to the Escherichia coli enzyme β-galactosidase.[11][12] This homologous relationship, along with the knowledge that glycosidases often perform hydrolysis catalyzed by two acidic residues, enabled the development of a mechanistic hypothesis. This hypothesis proposes that the two glutamic acid residues Glu540 and Glu451 are the nucleophilic and acidic residues, respectively, and that the tyrosine residue Tyr504 is also involved in catalysis.
In support of this hypothesis, experimental mutations in any of these three residues result in large decreases of enzymatic activity. Increased activity of an E451A mutant enzyme (where Glu451 is replaced with an alanine residue) after addition of azide is consistent with Glu451 as the acid/base residue.[13] Using analysis of labeled β-glucuronidase peptides after hydrolysis of a substrate that enters a very stable intermediate stage, researchers have determined that Glu540 is the nucleophilic residue.[14]
Though the particular type of nucleophilic substitution employed by β-glucuronidase is unclear, evidence for the mechanisms of their homologues in the glycosidase family suggests that these reactions are qualitatively SN2 reactions. The reactions proceed through a transition state with oxocarbenium ion characteristics. Initially, these mechanisms, because of this oxocarbenium characteristic of the transition state, were suggested to be SN1 reactions proceeding through a discrete oxocarbenium ion intermediate. However, more recent evidence suggests that these oxocarbenium ion states have lifetimes of 10 femtoseconds - 0.1 nanoseconds (similar to that of a bond vibration period). These lifetimes are too short to assign to a reaction intermediate. From this evidence, it appears that these reactions, while having an SN1 appearance due to the oxocarbenium ion characteristics of their transition states, must be qualitatively SN2 reactions.[2]
The specific activity of Tyr504 in the catalytic mechanism is unclear.[13] Through comparison to the structural data of the homologous enzyme xylanase, it has been suggested that Tyr504 of β-glucuronidase might stabilize the leaving nucleophile (Glu540) or modulate its activity.[15]
In addition to these residues, a conserved asparagine residue (Asn450) has been suggested to stabilize the substrate through the action of a hydrogen bond at the 2-hydroxyl group of the sugar substrate.[10][16]
.mw-parser-output .mod-gallerydisplay:table.mw-parser-output .mod-gallery-defaultbackground:transparent;margin-top:0.5em.mw-parser-output .mod-gallery-centermargin-left:auto;margin-right:auto.mw-parser-output .mod-gallery-leftfloat:left.mw-parser-output .mod-gallery-rightfloat:right.mw-parser-output .mod-gallery-nonefloat:none.mw-parser-output .mod-gallery-collapsiblewidth:100%.mw-parser-output .mod-gallery .titledisplay:table-row.mw-parser-output .mod-gallery .title>divdisplay:table-cell;text-align:center;font-weight:bold.mw-parser-output .mod-gallery .maindisplay:table-row.mw-parser-output .mod-gallery .main>divdisplay:table-cell.mw-parser-output .mod-gallery .captiondisplay:table-row;vertical-align:top.mw-parser-output .mod-gallery .caption>divdisplay:table-cell;display:block;font-size:94%;padding:0.mw-parser-output .mod-gallery .footerdisplay:table-row.mw-parser-output .mod-gallery .footer>divdisplay:table-cell;text-align:right;font-size:80%;line-height:1em.mw-parser-output .mod-gallery .gallerybox .thumb imgbackground:none.mw-parser-output .mod-gallery .bordered-images imgborder:solid #eee 1px.mw-parser-output .mod-gallery .whitebg imgbackground:#fff!important.mw-parser-output .mod-gallery .gallerybox divbackground:#fff!important
Repeating unit of the heparan sulfate substrate of β-glucuronidase

Surface depiction of active site pocket of β-glucuronidase with catalytic residues shown[1]
Mechanism of β-glucuronidase hydrolysis of a sugar substrate with high energy transition states showing oxocarbenium ion character depicted[14]
Potential stabilization of the nucleophilic residue Glu540 by Tyr504 in β-glucuronidase[15]
Predicted activity of the conserved Asn450 residue in stabilization of the β-glucuronidase sugar substrate[10][16]

Potential salt bridge between Glu352 and Arg216 in human beta-glucuronidase[1][17]
Sly syndrome
Deficiencies in β-glucuronidase result in the autosomal recessive inherited metabolic disease known as Sly syndrome or Mucopolysaccharidosis VII. A deficiency in this enzyme results in the build-up of non-hydrolyzed mucopolysaccharides in the patient. This disease can be extremely debilitating for the patient or can result in hydrops fetalis prior to birth. In addition, mental retardation, short stature, coarse facial features, spinal abnormalities, and enlargement of liver and spleen are observed in surviving patients.[5] This disease has been modeled in a strain of mice as well as a family of dogs.[18][19] More recently researchers have discovered a feline family that exhibits deficiencies in β-glucuronidase activity. The source of this reduction of activity has been identified as an E351K mutation (Glu351 is mutated to a lysine residue). Glu351 is conserved in mammalian species, which suggests an important function for this residue. Examination of the human X-ray crystal structure suggests that this residue (Glu352 in the human enzyme), which is buried deep within the TIM barrel domain, may be important for stabilization of the tertiary structure of the enzyme.[17] In the crystal structure, it appears that Arg216, a member of the jelly roll domain of the protein, forms a salt bridge with Glu352; therefore, Glu352 is likely involved in stabilizing the interaction between two different three-dimensional domains of the enzyme.[1]
Molecular applications: use as a reporter gene
In molecular biology, β-glucuronidase is used as a reporter gene to monitor gene expression in mammalian and plant cells. Monitoring β-glucuronidase activity through the use of a GUS assay allows determination of the spatial and temporal expression of the gene in question.[20]
- Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).[21]
See also
- Alpha-glucuronidase
- Glucuronosyl-disulfoglucosamine glucuronidase
- Glycyrrhizinate beta-glucuronidase
References
^ abcdef PDB: 1BHG; Jain S, Drendel WB, Chen ZW, Mathews FS, Sly WS, Grubb JH (April 1996). "Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs". Nat. Struct. Biol. 3 (4): 375–81. doi:10.1038/nsb0496-375. PMID 8599764..mw-parser-output cite.citationfont-style:inherit.mw-parser-output qquotes:"""""""'""'".mw-parser-output code.cs1-codecolor:inherit;background:inherit;border:inherit;padding:inherit.mw-parser-output .cs1-lock-free abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .cs1-lock-limited a,.mw-parser-output .cs1-lock-registration abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .cs1-lock-subscription abackground:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registrationcolor:#555.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration spanborder-bottom:1px dotted;cursor:help.mw-parser-output .cs1-hidden-errordisplay:none;font-size:100%.mw-parser-output .cs1-visible-errorfont-size:100%.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-formatfont-size:95%.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-leftpadding-left:0.2em.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-rightpadding-right:0.2em
^ abc Sinnott, Michael, ed. (1998). Comprehensive Biological Catalysis. 1. Manchester, UK: Academic Press. pp. 119–138. ISBN 0-12-646864-8.CS1 maint: Extra text: authors list (link)
^ McCarter JD, Withers SG (1994). "Mechanisms of enzymatic glycoside hydrolysis". Curr Opin Struct Biol. 4 (6): 885–892. doi:10.1016/0959-440X(94)90271-2. PMID 7712292.
^ Sinnott ML (1990). "Catalytic mechanisms of enymatic glycosyl transfer". Chem Rev. 90: 1171–1202. doi:10.1021/cr00105a006.
^ ab Nyhan, William L.; Bruce Barshop; Pinar Ozand (2005). Atlas of Metabolic Diseases (2 ed.). London, UK: Hodder Arnold. pp. 501–503, 546–550. ISBN 0-340-80970-1.
^ Oshima A, Kyle JW, Miller RD, Hoffmann JW, Powell PP, Grubb JH, Sly WS, Tropak M, Guise KS, Gravel RA (Mar 1987). "Cloning, sequencing, and expression of cDNA for human beta-glucuronidase". Proc Natl Acad Sci U S A. 84 (3): 685–9. doi:10.1073/pnas.84.3.685. PMC 304280. PMID 3468507.
^ "Entrez Gene: GUSB glucuronidase, beta".
^ Islam MR, Grubb JH, Sly WS (1993). "C-terminal processing of human beta-glucuronidase. The propeptide is required for full expression of catalytic activity, intracellular retention, and proper phosphorylation". J Biol Chem. 268 (16): 12193–12198. PMID 8226771.
^ Shipley JM, Grubb JH, Sly WS (1993). "The role of glycosylation and phosphorylation in the expression of active human β-glucuronidase". J Biol Chem. 268 (16): 12193–12198. PMID 8505339.
^ abc Kim HW, Mino K, Ishikawa K (December 2008). "Crystallization and preliminary X-ray analysis of endoglucanase from Pyrococcus horikoshii". Acta Crystallographica Section F. 64 (Pt 12): 1169–71. doi:10.1107/S1744309108036919. PMC 2593689. PMID 19052378.
^ Henrissat B, Bairoch A (1993). "New families in the classification of glycosyl hydrolases". Biochem J. 293: 781–788. doi:10.1042/bj2930781. PMC 1134435. PMID 8352747.
^ Henrissat B (1991). "A classification of glycosyl hydrolases based on amino-acid sequence similarities". Biochem J. 280: 309–316. doi:10.1042/bj2800309. PMC 1130547. PMID 1747104.
^ ab Islam MR, Tomatsu S, Shah GN, Grubb JH, Sanjeev J, Sly WS (1999). "Active site residues of human β-glucuronidase. Evidence for Glu(540) as the nucleophile and Glu(451) as the acid-base residue". J Biol Chem. 274 (33): 23451–23455. doi:10.1074/jbc.274.33.23451. PMID 10438523.
^ ab Wong AW, He S, Grubb JH, Sly WS, Withers SG (1996). "Identification of Glu-540 as the catalytic nucleophile of human β-glucuronidase using electrospray mass spectrometry". J Biol Chem. 273 (51): 34057–34062. doi:10.1074/jbc.273.51.34057. PMID 9852062.
^ ab "EzCatDB: T00066". EzCatDB: A Database of Catalytic Mechanisms. Archived from the original on 2009-06-17. Retrieved 2008-12-12.
^ ab Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G (1995). "Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases". Proc Natl Acad Sci USA. 92 (15): 7090–7094. doi:10.1073/pnas.92.15.7090. PMC 41477. PMID 7624375.
^ ab Fyfe JC, Kurzhals RL, Lassaline ME, Henthorn PS, Alur PR, Wang P, Wolfe JH, Giger U, Haskins ME, Patterson DF, Sun H, Jain S, Yuhki N (1999). "Molecular basis of feline β-glucuronidase deficiency: An animal model of mucopolysaccharidosis VII". Genomics. 58 (2): 121–128. doi:10.1006/geno.1999.5825. PMID 10366443.
^ Birkenmeier EH; Davisson MT; Beamer WG; Ganschow RE; Vogler CA; Gwynn B; Lyford KA; Maltais LM; Wawrzyniak CJ (1989). "Murine mucopolysaccaaridosis type VII: Characterization of a mouse with beta-glucuronidase deficiency". J Clin Invest. 83 (4): 1258–1266. doi:10.1172/JCI114010. PMC 303816. PMID 2495302.
^ Haskins ME, Desnick RJ, DiFerrante N, Jezyk PF, Patterson DF (1984). "β-glucuronidase deficiency in a dog: A model of human mucopolysaccharidosis VII". Pediatr Res. 18 (10): 980–984. doi:10.1203/00006450-198410000-00014. PMID 6436780.
^ Marathe SV, McEwen JE (February 1995). "Vectors with the gus reporter gene for identifying and quantitating promoter regions in Saccharomyces cerevisiae". Gene. 154 (1): 105–7. doi:10.1016/0378-1119(94)00845-J. PMID 7867935.
^ Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (October 2004). "UCSF Chimera--a visualization system for exploratory research and analysis". J Comput Chem. 25 (13): 1605–12. doi:10.1002/jcc.20084. PMID 15264254.
Further reading
.mw-parser-output .refbeginfont-size:90%;margin-bottom:0.5em.mw-parser-output .refbegin-hanging-indents>ullist-style-type:none;margin-left:0.mw-parser-output .refbegin-hanging-indents>ul>li,.mw-parser-output .refbegin-hanging-indents>dl>ddmargin-left:0;padding-left:3.2em;text-indent:-3.2em;list-style:none.mw-parser-output .refbegin-100font-size:100%
George J. (2008). "Elevated serum beta-glucuronidase reflects hepatic lysosomal fragility following toxic liver injury in rats". Biochem Cell Biol. 86 (3): 235–243. doi:10.1139/O08-038. PMID 18523484.
Bell CE, Sly WS, Brot FE (1977). "Human beta-glucuronidase deficiency mucopolysaccharidosis: identification of cross-reactive antigen in cultured fibroblasts of deficient patients by enzyme immunoassay". J. Clin. Invest. 59 (1): 97–105. doi:10.1172/JCI108627. PMC 333336. PMID 401508.
Tanaka J, Gasa S, Sakurada K, et al. (1992). "Characterization of the subunits and sugar moiety of human placental and leukemic beta-glucuronidase". Biol. Chem. Hoppe-Seyler. 373 (1): 57–62. doi:10.1515/bchm3.1992.373.1.57. PMID 1311180.
Wolfe JH, Sands MS, Barker JE, et al. (1993). "Reversal of pathology in murine mucopolysaccharidosis type VII by somatic cell gene transfer". Nature. 360 (6406): 749–53. doi:10.1038/360749a0. PMID 1465145.
Tomatsu S, Fukuda S, Sukegawa K, et al. (1991). "Mucopolysaccharidosis type VII: characterization of mutations and molecular heterogeneity". Am. J. Hum. Genet. 48 (1): 89–96. PMC 1682743. PMID 1702266.
Shipley JM, Miller RD, Wu BM, et al. (1991). "Analysis of the 5' flanking region of the human beta-glucuronidase gene". Genomics. 10 (4): 1009–18. doi:10.1016/0888-7543(91)90192-H. PMID 1916806.
Ono M, Taniguchi N, Makita A, et al. (1988). "Phosphorylation of beta-glucuronidases from human normal liver and hepatoma by cAMP-dependent protein kinase". J. Biol. Chem. 263 (12): 5884–9. PMID 2833520.
Guise KS, Korneluk RG, Waye J, et al. (1985). "Isolation and expression in Escherichia coli of a cDNA clone encoding human beta-glucuronidase". Gene. 34 (1): 105–10. doi:10.1016/0378-1119(85)90300-2. PMID 3924735.
Ho YC, Ho LH, Ho KJ (1985). "Human hepatic beta-glucuronidase: an enzyme kinetic study". Enzyme. 33 (1): 9–17. PMID 3987656.
Shipley JM, Klinkenberg M, Wu BM, et al. (1993). "Mutational analysis of a patient with mucopolysaccharidosis type VII, and identification of pseudogenes". Am. J. Hum. Genet. 52 (3): 517–26. PMC 1682147. PMID 7680524.
Vervoort R, Lissens W, Liebaers I (1994). "Molecular analysis of a patient with hydrops fetalis caused by beta-glucuronidase deficiency, and evidence for additional pseudogenes". Hum. Mutat. 2 (6): 443–5. doi:10.1002/humu.1380020604. PMID 8111412.
Wu BM, Sly WS (1994). "Mutational studies in a patient with the hydrops fetalis form of mucopolysaccharidosis type VII". Hum. Mutat. 2 (6): 446–57. doi:10.1002/humu.1380020605. PMID 8111413.
Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
Moullier P, Bohl D, Heard JM, Danos O (1993). "Correction of lysosomal storage in the liver and spleen of MPS VII mice by implantation of genetically modified skin fibroblasts". Nat. Genet. 4 (2): 154–9. doi:10.1038/ng0693-154. PMID 8348154.
Shipley JM, Grubb JH, Sly WS (1993). "The role of glycosylation and phosphorylation in the expression of active human beta-glucuronidase". J. Biol. Chem. 268 (16): 12193–8. PMID 8505339.
Nishimura Y, Kato K, Himeno M (1996). "Biochemical characterization of liver microsomal, Golgi, lysosomal, and serum beta-glucuronidases in dibutyl phosphate-treated rats". J. Biochem. 118 (1): 56–66. PMID 8537326.
Jain S, Drendel WB, Chen ZW, et al. (1996). "Structure of human beta-glucuronidase reveals candidate lysosomal targeting and active-site motifs". Nat. Struct. Biol. 3 (4): 375–81. doi:10.1038/nsb0496-375. PMID 8599764.
Vervoort R, Islam MR, Sly WS, et al. (1996). "Molecular analysis of patients with beta-glucuronidase deficiency presenting as hydrops fetalis or as early mucopolysaccharidosis VII". Am. J. Hum. Genet. 58 (3): 457–71. PMC 1914559. PMID 8644704.
Bonaldo MF, Lennon G, Soares MB (1997). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Res. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
Dentino AR, Raj PA, De Nardin E (1997). "Subtle differences between human and rabbit neutrophil receptors shown by the secretagogue activity of constrained formyl peptides". Arch. Biochem. Biophys. 337 (2): 267–74. doi:10.1006/abbi.1996.9791. PMID 9016822.
External links
Glucuronidase at the US National Library of Medicine Medical Subject Headings (MeSH)
Updated research on reporter glucuronidase and other reporters from Reportergene
Database of Catalytic Mechanism Research and other information on beta-glucuronidase